For an upcoming workshop I was looking for an interesting mix of genomics and machine learning to show off Azure capabilities. My MS Genomics colleague Erdal Cosgun recently showed me his workshop material, which uses MS Genomics to predict variants and Azure Machine Learning Studio to predict (fake) phenotypes for the samples (in a fancy no-code GUI). I liked the idea and also wanted to try out something new: Azure AutoML, a framework that originated in Microsoft Research.
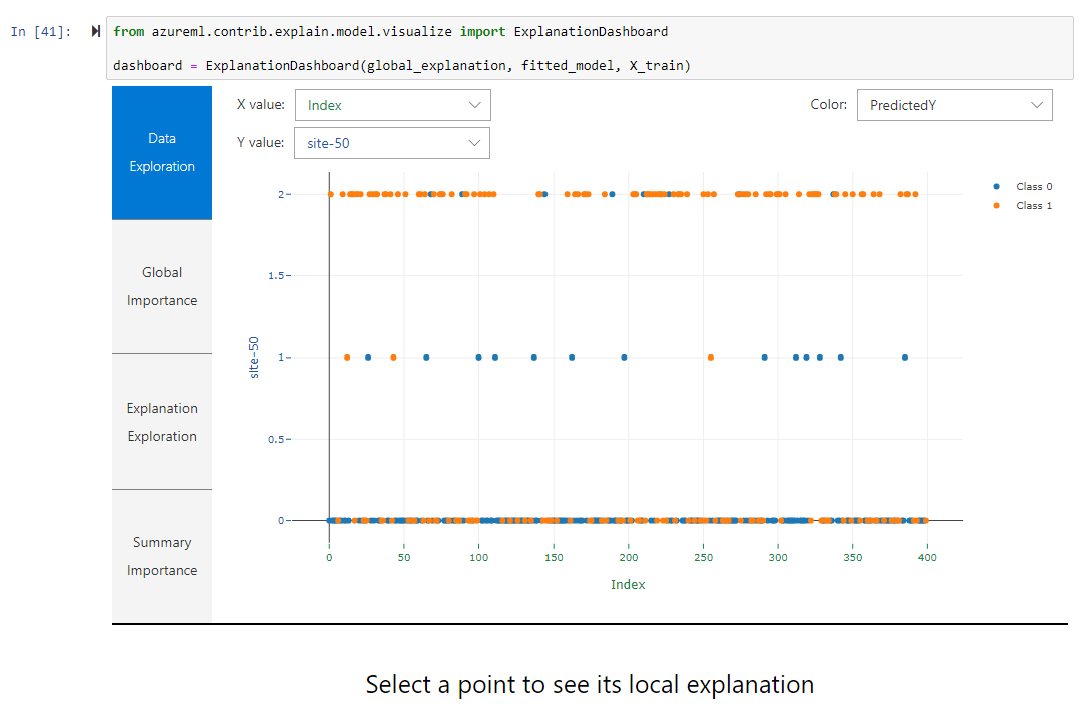
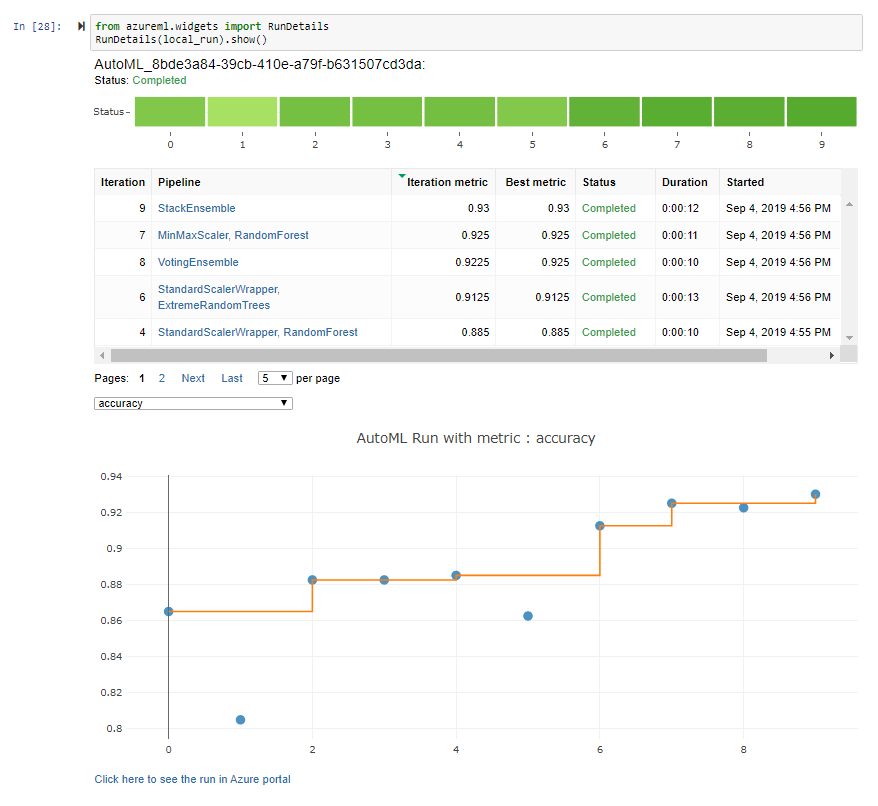
AutoML basically takes care of the time-consuming, iterative tasks of ML model development. It trains & tunes a model, using the target metric you specify and iterates through multiple ML algorithms. Each iteration produces a model with a training score and everything is logged as an “Experiment” in the Azure portal. You could say it’s a recommender system for ML pipelines that allows me to employ a range of ML techniques and practices without having to be an expert. It further has preprocessing (e.g. data imputation, encoding, embedding) and interpretation capabilities that can explain a models behaviour. The latter shows me for example which features where the most important ones. That follows the transparency principle, one of the six Microsoft AI principles (ever asked other cloud providers regarding theirs?).
Initially I had planned to just use the AutoML Visual Interface in the Azure Portal, so that I (and later the workshop participants) would not have to write any code. Unfortunately, the variant call format (vcf) is not really suitable for this. It usually has thousands of columns and the visual interface tries to preview all columns/features, which of course doesn’t work at that scale. So instead I ran the analysis on an Azure ML Notebook VM using the AutoML Python API.
I also decided to create my own input data: faked variant calls from a number of individuals and a limited number of sites, so that I could complete any analysis just on the Notebook VM. I made multiple sites “causal” and furthermore introduced a gender bias. The input data creation is scripted in the notebook itself and can be easily rerun to create new data with different features. The notebook shows how, with a few lines of Python, the AutoML system produces a highly performant model that picked the correct causal sites and gender as decisive factors for predicting a phenotype. The input data is obviously not fully representative of real data, but the capabilities of the AutoML platform are nicely demonstrated. This was fun and I hope the workshop participants will like this exercise.
The notebook covers:
- Input data generation
- Running AutoML
- Predicting outcome and plotting a confusion matrix
- Interpretation of the model
You can find the notebook here as HTML rendered Notebook or on Github as Jupyter Notebook. Unfortunately neither of these shows the AutoML widgets imported from the Azure Portal, so I added screenshots below:
AutoML Run Details
Explanation Dashboard